Up-to-date Nociceptor Neurons Meta-Analysis
Supplementary ressource for:
Crosson T, Roversi K, Balood M, Othman R, Ahmadi M, Wang JC, Pereira PJS, Couture R, Latini AS, Prediger RDS, Rangachari M, Seehus CR, Foster SL, Talbot S*. Profiling of how nociceptor neurons detect danger; new and old foes. Journal of Internal Medicine. 2019
Click the download dataset link to upload and explore the complete meta-analysis :
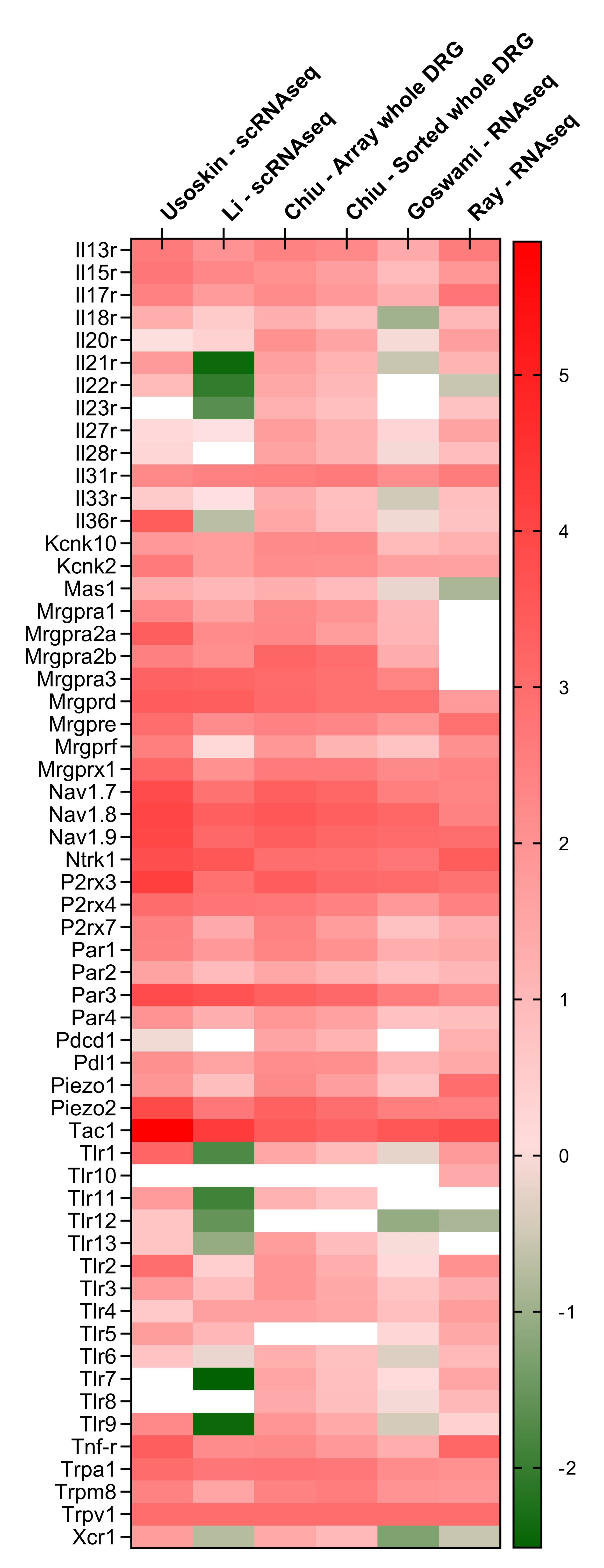
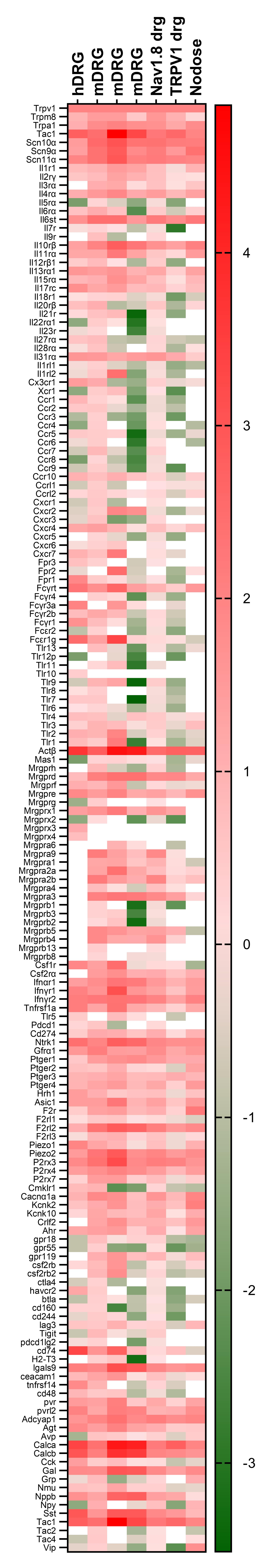
Nociceptors have been reported in signal studies to express a variety of “danger detecting” receptors, but as of yet there has not been a cumulative summary of these findings. Therefore, we generated a meta-analysis of four naïve mice and one human dorsal root ganglia profiling datasets.
First, Chiu [52] and colleagues’ microarray (28000 coding and 7000 non-coding transcripts) profiled whole and FACS-sorted NaV1.8+ DRGs neurons. The cells used were pooled from 7–20-week-old age-matched male and female cervical (C1–C8), thoracic (T1–T13), and lumbar (L1–L6) ganglia. Relative gene expression was reported as robust multiarray average (RMA). Ray et al., [53] RNA-sequenced L2 DRG neurons from three Caucasian women aged 30-60 years and reported expression levels as transcripts per kilobase million (TPM). Goswami et al., [54] RNA-sequenced FACS-sorted TRPV1+ L3-L5 DRG neurons dissected from 3 groups of ten 8-week-old male and female C57Bl6 mice and reported expression levels as reads per kilobase per million mapped reads (RPKM). Usoskin and colleague’s [55] RNA-sequenced (1.14 million reads were mapped to 3,574 ± 2,010 genes/cell) single neurons from L4–L6 lumbar DRGs harvested from 6–8-week-old male and female adult mice. The 799 cells sequenced were randomly picked, of which 622 cells were classified as neurons, 68 cells had an ambiguous assignment, and 109 cells were non-neuronal. Data were reported as numbers of read per million (RPM), which, for comparison, were transformed as RPKM (Figure 1) Li and colleagues [56] performed high-coverage single-cell RNA sequencing (10950 ± 1 218 genes/neuron) of 64 IB4+ neurons, 69 IB4- small neurons and 64 large neurons collected from nineteen 8-10 weeks male C57BL/6 mice and data expressed as RPKM.
Dataset referenced:
52 Chiu IM, Barrett LB, Williams EK, et al. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. eLife 2014; 3.
53 Ray P, Torck A, Quigley L, et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018; 159: 1325-45.
54 Goswami SC, Mishra SK, Maric D, et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. The journal of pain : official journal of the American Pain Society 2014; 15: 1338-59.
55 Usoskin D, Furlan A, Islam S, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nature neuroscience 2015; 18: 145-53.
56 Li C, Wang S, Chen Y, Zhang X. Somatosensory Neuron Typing with High-Coverage Single-Cell RNA Sequencing and Functional Analysis. Neurosci Bull 2018; 34: 200-7.